PPM1D and the DNA Damage Response in Hematopoietic Cells
PPM1D Mutations Are Common after Chemotherapy
Numerous studies have now confirmed that individuals with clonal hematopoiesis, myelodysplastic syndrome (MDS), or acute myeloid leukemia (AML) who have previously been exposed to chemotherapy or radiation are significantly more likely to have a mutation in a gene involved in the DNA damage response (DDR). Among this group of genes, PPM1D is the most-commonly mutated, with recurrent truncating mutations occurring in the c-terminus of the protein (1).
PPM1D Truncations Impair Degradation
We have previously shown that the c-terminus of PPM1D encodes for a degradation signal by the proteasome. Therefore, loss of the c-terminus of PPM1D via truncating mutations impair proteasomal degradation and increase intracellular levels of the protein. These elevated levels in turn suppress the DNA damage response and p53 activation allowing cells harboring these truncating mutations to be more resistant to chemotherapy relative to non-mutant cells (2).
PPM1D as a Therapeutic Target in Cancer
We have now developed numerous new tools to interrogate these findings and are now working to more deeply explore numerous aspects of PPM1D biology, ranging from fundamental structure-functional studies of PPM1D to therapeutic studies in vivo. We believe that these lines of inquiry are required to understand the biology of PPM1D, the stem cell response to genotoxic stress, and the potential of PPM1D as a drug target in cancer.
Three Key Questions:
1) How do mutations in PPM1D affect the protein and why are they selected for in stem cells that are exposed the DNA damaging agents?
2) What do the emergence and effects of these PPM1D mutations teach us more generally about how normal and cancerous stem cells deal the genotoxic stress?
3) Is PPM1D a potential therapeutic target in myelodysplastic syndrome, acute myeloid leukemia, and other tumors?
Selected References
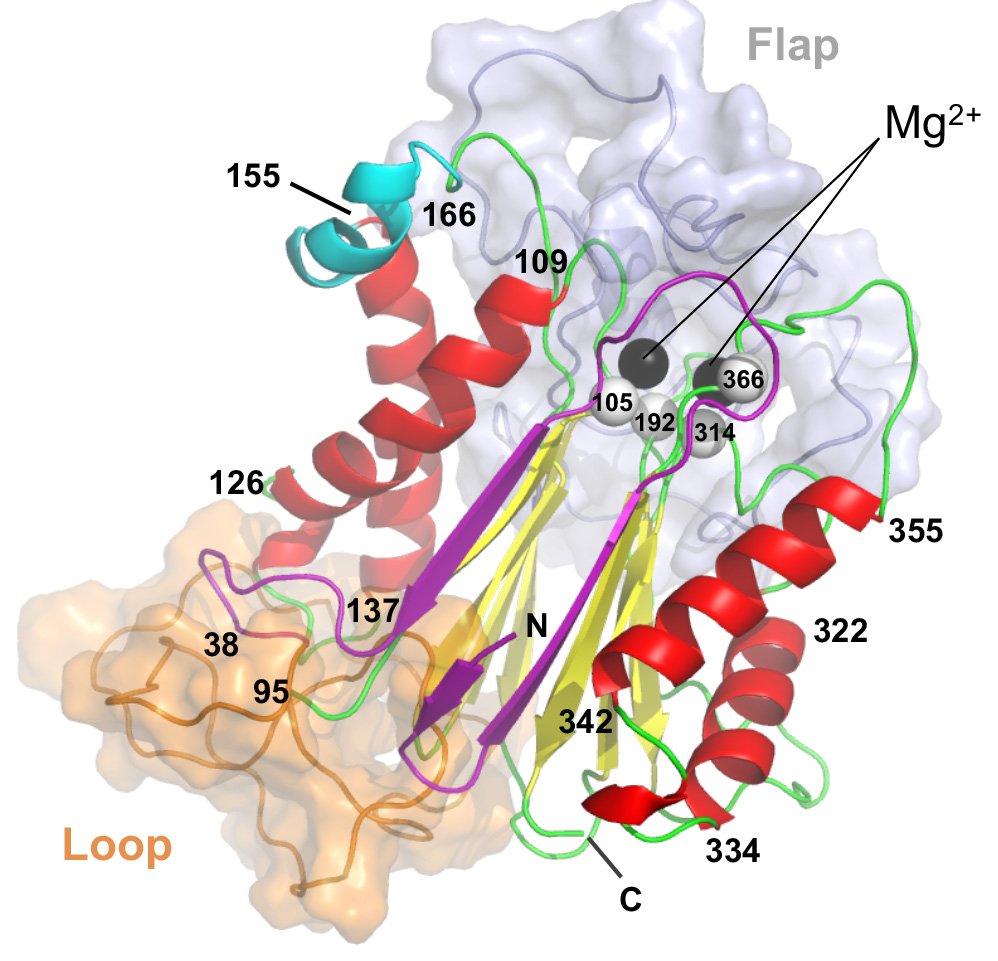
(1) Allosteric inhibition of PPM1D serine/threonine phosphatase via an altered conformational state (Pubmed)
Miller PG*, Sathappa M*, Moroco JA*, Jiang W, Qian Y, Iqbal S, Guo Q, Giacomelli AO, Shaw S, Vernier C, Bajrami B, Yang X, Raffier C, Sperling AS, Gibson CJ, Kahn J, Jin C, Ranaghan M, Caliman A, Brousseau M, Fischer ES, Lintner R, Piccioni F, Campbell AJ, Root DE, Garvie CW, Ebert BL.
Nature Communications 2022. June 30;13(1):3778
*Authors Contributed Equally to This Work
(2) Clonal Hematopoiesis in Patients Receiving Chimeric Antigen Receptor T-Cell Therapy (Pubmed)
Miller PG*, Sperling AS*, Brea EJ, Leick MB, Fell GG, Jan M, Gohil SH, Tai Y, Munshi NC, Wu CJ, Neuberg DS, Maus MV, Jacobson C, Gibson CJ, Ebert BL.
Blood Advances. 2021. Aug 10;5(15):2982-2986
*Authors Contributed Equally to This Work
(3) A dominant-negative effect drives selection of TP53 missense mutations in myeloid malignancies (Pubmed)
Boettcher S, Miller PG, Sharma R, McConkey M, Leventhal M, Krivtsov AV, Giacomelli AO, Wong W, Kim J, Chao S, Kurppa KJ, Yang X, Milenkowic K, Piccioni F, Root DE, Rücker FG, Flamand Y, Neuberg D, Lindsley RC, Jänne PA, Hahn WC, Jacks T, Döhner H, Armstrong SA, Ebert BL.
Science. 2019 Aug 9;365(6453):599-604.
(4) PPM1D truncating mutations confer resistance to chemotherapy and sensitivity to PPM1D inhibition in hematopoietic cells (Pubmed)
Kahn JD, Miller PG, Silver AJ, Sellar RS, Bhatt S, Gibson C, McConkey M, Adams D, Mar B, Mertins P, Fereshetian S, Krug K, Zhu H, Letai A, Carr SA, Doench J, Jaiswal S, Ebert BL.
Blood. 2018 Sep 13;132(11):1095-1105.